The Bioinformatics and Intelligent Medicine Team published one research paper in
Cell Reports Methods.
October 28,2024 Editor:Centre for Bioinformatics and Intelligent
Medicine
Recently, the research collaboration among the Bioinformatics & Intelligent Medicine team,
Southern Medical University
Zhujiang Hospital, Tianjin Medical University Cancer Hospital, etc. has resulted in the
publication titled “Precise
detection of cell-type-specific domains in spatial transcriptomics” in the journal
Cell Reports Methods.

In clinical practice, accurately dissecting the spatial heterogeneity of cell types within
tissues is crucial for
understanding disease mechanisms, assessing prognostic risks, and developing personalized
treatment strategies. However,
traditional spatial domain analysis methods face significant challenges in spatial
transcriptomics (SRT) research,
particularly when detecting the spatial localization of low-proportion cell types. These cells
often overlap with or are
located within other high-proportion cell types, making it difficult to identify them
accurately.
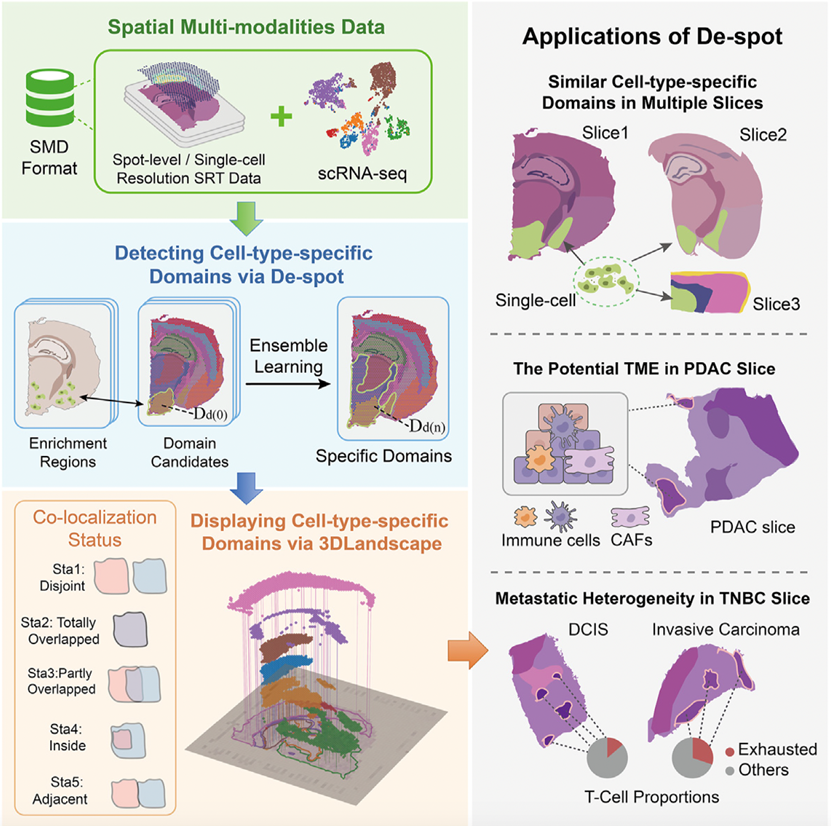
To the challenges above, the research team proposed the novel concept of "cell-type-specific
domains", which are further
expected to be spatially heterogeneous domains where the proportions of given cell types are
significantly higher than
other domains. Compared to conventional spatial domains, this concept can better characterize
the anatomical regions
where low-proportion cell types reside in spatial transcriptomics. Further, the research team
developed De-spot that
enables precise detection of cell-type-specific domains in both spot-level and single-cell SRT
slices. De-spot bridges
the gap between cell-type proportions and spatial domains by integrating potential candidates,
inferring enrichment
regions of cell-type proportions, and mapping enrichment regions to spatial domains. De-spot
takes advantage of
segmentation and deconvolution simultaneously, making it more effective in selecting
cell-type-specific domains as ROIs
without manual fine-tuning, including dealing with low-proportion cell types.
Furthermore, the research team also developed a new spatial multi-modal data format (SMD) with
extensible interfaces,
which are compatible with multiple SRT platforms, greatly enhancing its versatility and
practicality. To visually
present the cell type-specific domains, the research team also developed the 3D Landscape,
enabling researchers to gain
a more intuitive understanding of the distribution and interactions of cell type-specific
domains in space.
De-spot not only addresses the limitations of current computational methods in detecting cell
type-specific spatial
domains but also provides a new perspective for exploring complex biological processes such as
the tumor
microenvironment (TME).
Experimental evaluation showed that De-spot discovered the co-localizations between
cancer-associated fibroblasts and
immune-related cells that indicate potential tumor microenvironment (TME) domains in given
slices, which were obscured
by previous computational methods. The research team further elucidated the identified domains
and found that Srgn may
be a critical TME marker in SRT slices. By deciphering T cell-specific domains in breast cancer
tissues, De-spot also
revealed that the proportions of exhausted T cells were significantly increased in invasive vs.
ductal carcinoma. By
precisely identifying cell-type-specific domains, De-spot offers new insights into disease
mechanisms and the
optimization of treatment strategies in clinical research.