The Center for Bioinformatics and Intelligent Medicine team published a paper in Briefings in Bioinformatics
May 03,2023 Editor:Centre for Bioinformatics and Intelligent Medicine
The lab team published “LPAD: using network construction and label propagation to detect topologically associating domains from Hi-C data” in Briefings in Bioinformatics.
The high-throughput Chromosome Conformation Capture (Hi-C) sequencing technology has brought significant advancements in the study of the three-dimensional spatial organization of the genome, offering a new paradigm for understanding long-range chromosomal regulatory mechanisms. Hi-C technology has revealed the highly folded nature of chromosomes in three-dimensional space, forming a structure known as topologically associated domains (TADs), which are considered as the fundamental regulatory units in chromosomal spatial organization. TADs play a crucial role in modulating gene expression by suppressing it within specific boundaries and establishing insulation between the regulatory boundaries, which holds significant implications for studying gene expression, such as the expression of tumor suppressor genes.
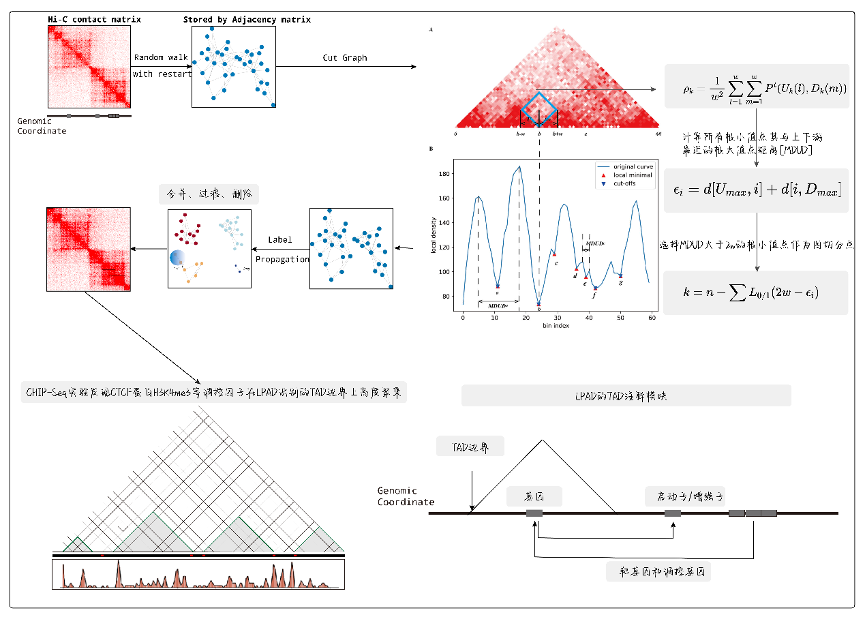
LPAD is a novel algorithm designed to identify topologically associated domain (TAD) structures from the Hi-C contact matrix. Considering that TAD structures arise from highly interacting and deeply folded spatial configurations within a contiguous chromosomal region, LPAD introduces a novel approach utilizing a random restart random walk algorithm to extract global information, including the correlation between contact points and their neighboring contact points, thereby constructing an undirected weighted graph representing chromosomal contact patterns. LPAD employs a linear-model method to partition the global, large-scale network graph into smaller subgraphs and designs an improved label propagation algorithm to identify community structures within the undirected graph. Then, LPAD classifies the community structures into TADs, TAD boundaries, and TAD gaps, representing distinct spatial configurations. Through experimental validation, including CHIP-Seq, LPAD demonstrates significant advantages in terms of accuracy and quality in TAD identification.