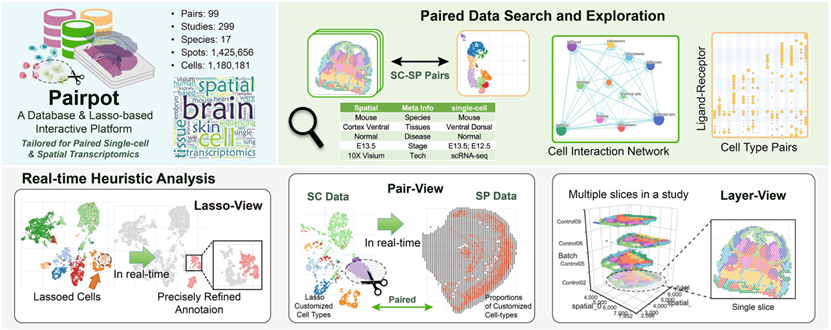
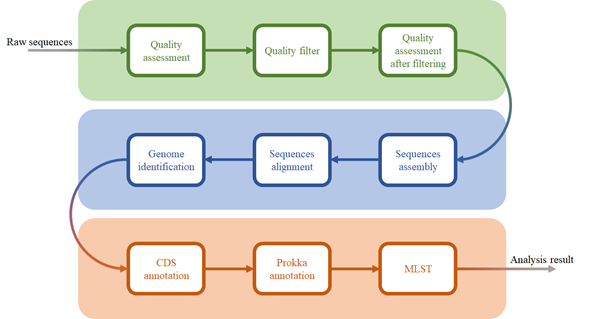
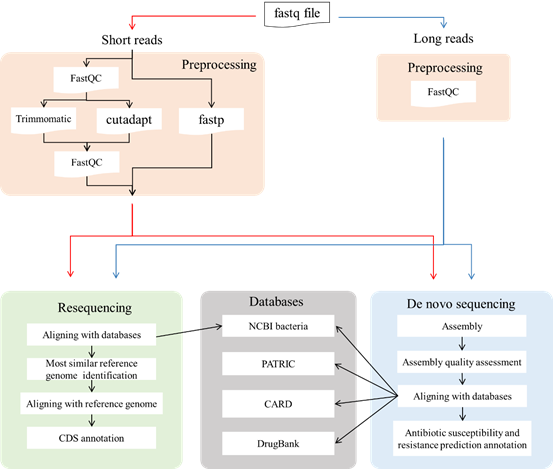
Pairpot
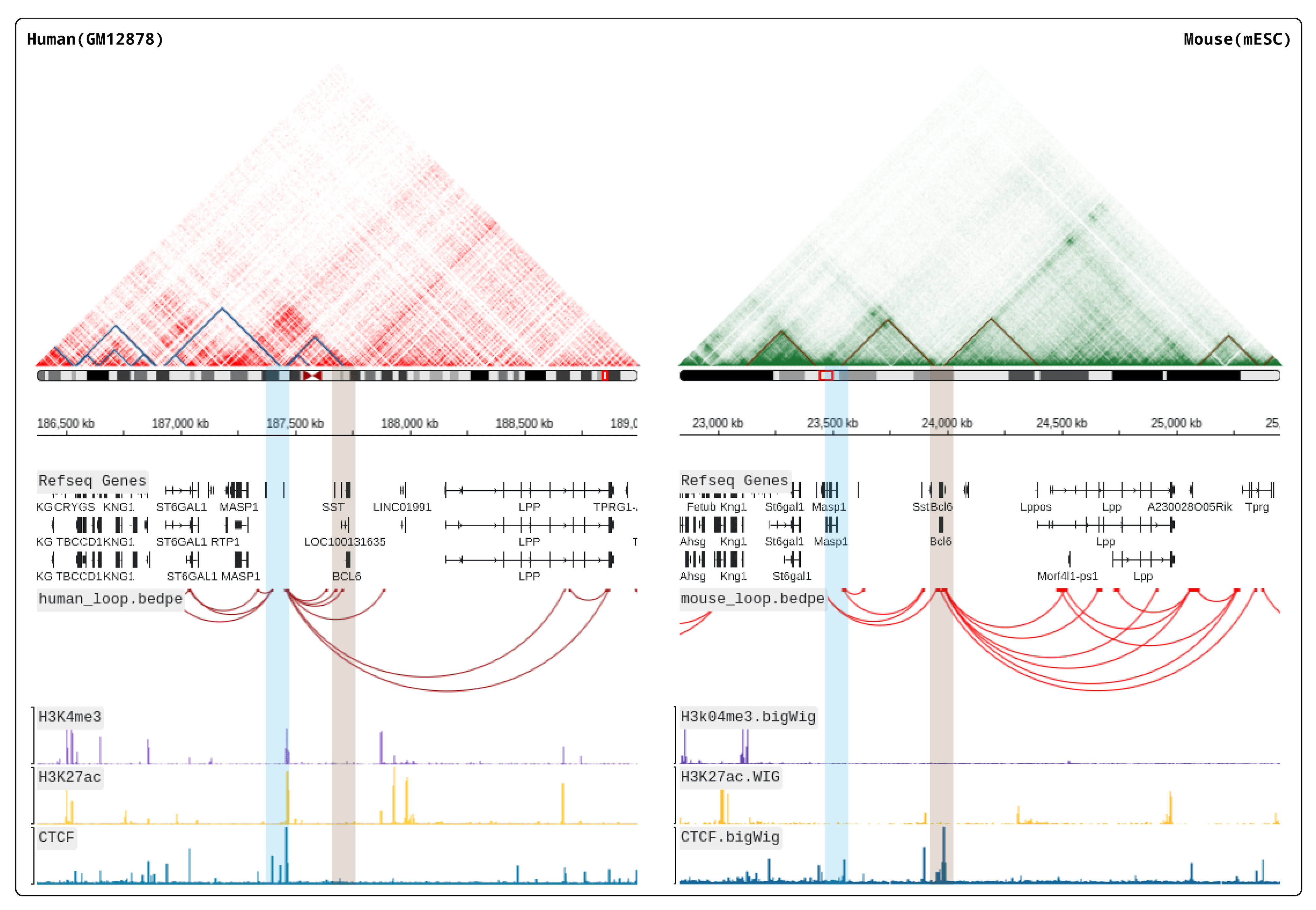
Pairpot is a database tailored for paired single-cell and SRT data with real-time heuristic analysis. It constructs the curated pairs by integrating multiple slices and establishing potential associations between single-cell and SRT data. On this basis, Pairpot adopts semi-supervised learning that enables real-time heuristic analysis where Lasso-View refines the user-selected SRT domains within milliseconds, Pair-View infers cell proportions of spots based on user-selected cell types in real-time and Layer-View displays SRT slices using a 3D hierarchical layout.